|
Part 1: Extracting Gene Names & DNA (3)
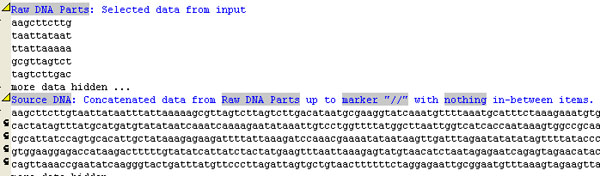
Now you are in the Convert Data panel again. We want to concatenate the dna fragments from the different columns into one long sequence so that we can manipulate it later. Click on Insert and click the the rule “Concatenante Data items from other rules”. Give the rule a meaningful name such as “Source DNA”. Click on the various grey highlighted regions to modify the rule to say the following:
Concatenanted data from Raw DNA Sequence up to marker "//" with nothing between items.
Note, you didn't do the concatenation with level in this tutorial since that would create a synchronization problems if you are processing more than one input files. Level-based concatenation will not be completed at the end of each BAC sequence but will be at the beginning of the next BAC sequence or the end of the input, which would be incorrect for this tutorial since we have multiple subsequences to be extracted from each BAC sequence. In general, always prefer to use an end maker to identify the end of a concatenation rather than using levels for complex tasks like this one. The // maker would have been selected along with the DNA fragments in the previous step. If that is not the case, the rule should be sent back to the Input panel and edited..
We should now have one big long DNA sequence, as in the following screenshot:
 
|